Genes
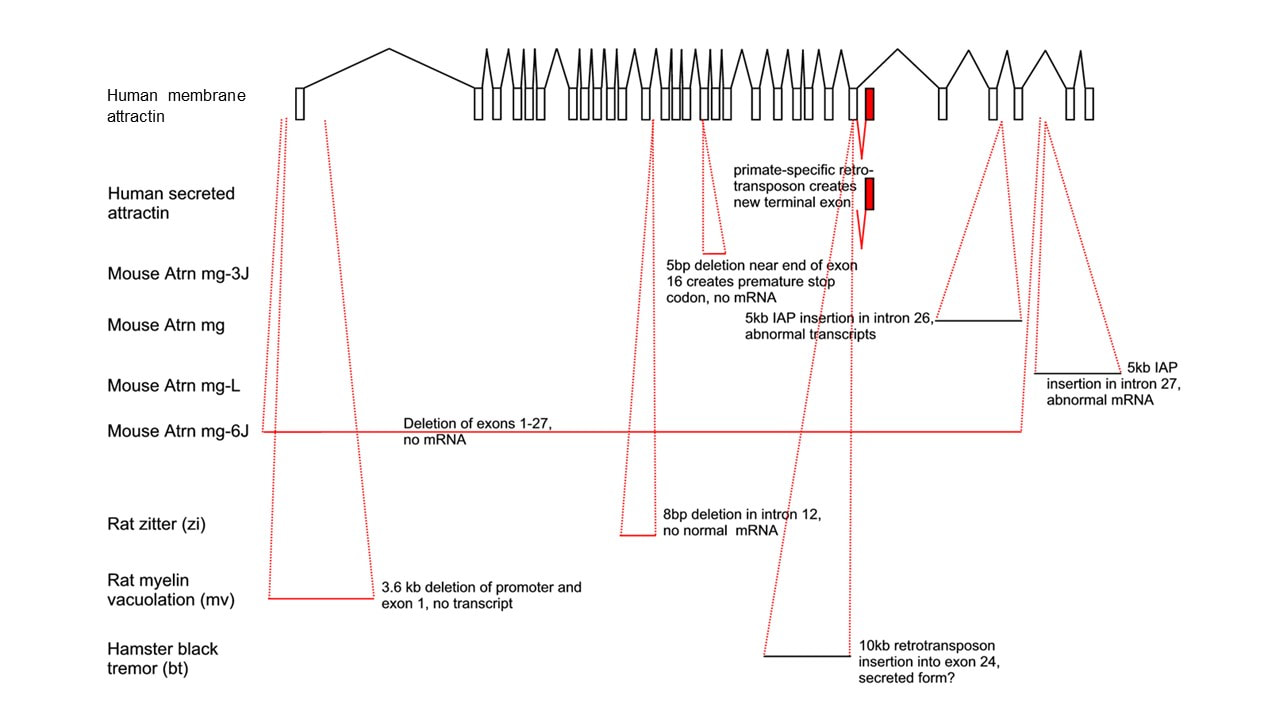
Although several isoforms of attractin have been shown or predicted to exist, evidence at the mRNA and protein level suggests that the full transmembrane transcript is the main homolog shared amongst metazoan organisms. A secreted form arising by alternative splicing has been shown to exist in higher primates including chimpanzees, gorillas and humans. The genomic organization consists of 30 potential exons (located in the 20p13 region in humans) where the common transmembrane form arises from sequential transcription of exons 1-24 followed by exons 26-30 where exon 26 encodes a transmembrane region. The alternate secreted form results from the exon 1-24 transcript reading straight through into exon 25 that encodes a stop codon and poly(A) signal.
Interestingly, exon 25 results from an ancient LINE-1 element insertion in primates, and the read-through to generate this alternate transcript is associated with cell activation in immune cells. In contrast, differentiation of neurons from human cortical neuron precursors results in a strong damping down of the secreted form transcription. The epigenetic mechanisms affecting transcription through the exon 24-25-26 region have not yet been elucidated.
The figure below outlines the main gene transcripts and the mutant forms expressed in several animal models. In these mutant models, common pathologies are spongiform vacuolation and hypomyelination with consequences often including tremor.
Interestingly, exon 25 results from an ancient LINE-1 element insertion in primates, and the read-through to generate this alternate transcript is associated with cell activation in immune cells. In contrast, differentiation of neurons from human cortical neuron precursors results in a strong damping down of the secreted form transcription. The epigenetic mechanisms affecting transcription through the exon 24-25-26 region have not yet been elucidated.
The figure below outlines the main gene transcripts and the mutant forms expressed in several animal models. In these mutant models, common pathologies are spongiform vacuolation and hypomyelination with consequences often including tremor.